Introduction¶
La sabiduría no vale la pena si no es posible servirse de ella para inventar una nueva manera de preparar los garbanzos.
[Wisdom isn’t worth anything if you can’t use it to come up with a new way to cook garbanzos.]
—Gabriel García Márquez, A wise Catalan in “Cien años de soledad”
The goal of PyTables is to enable the end user to manipulate easily data tables and array objects in a hierarchical structure. The foundation of the underlying hierarchical data organization is the excellent HDF5 library (see [HDGF1]).
It should be noted that this package is not intended to serve as a complete wrapper for the entire HDF5 API, but only to provide a flexible, very pythonic tool to deal with (arbitrarily) large amounts of data (typically bigger than available memory) in tables and arrays organized in a hierarchical and persistent disk storage structure.
A table is defined as a collection of records whose values are stored in fixed-length fields. All records have the same structure and all values in each field have the same data type. The terms fixed-length and strict data types may seem to be a strange requirement for an interpreted language like Python, but they serve a useful function if the goal is to save very large quantities of data (such as is generated by many data acquisition systems, Internet services or scientific applications, for example) in an efficient manner that reduces demand on CPU time and I/O.
In order to emulate in Python records mapped to HDF5 C structs PyTables implements a special class so as to easily define all its fields and other properties. PyTables also provides a powerful interface to mine data in tables. Records in tables are also known in the HDF5 naming scheme as compound data types.
For example, you can define arbitrary tables in Python simply by declaring a class with named fields and type information, such as in the following example:
class Particle(IsDescription):
name = StringCol(16) # 16-character String
idnumber = Int64Col() # signed 64-bit integer
ADCcount = UInt16Col() # unsigned short integer
TDCcount = UInt8Col() # unsigned byte
grid_i = Int32Col() # integer
grid_j = Int32Col() # integer
# A sub-structure (nested data-type)
class Properties(IsDescription):
pressure = Float32Col(shape=(2,3)) # 2-D float array (single-precision)
energy = Float64Col(shape=(2,3,4)) # 3-D float array (double-precision)
You then pass this class to the table constructor, fill its rows with your values, and save (arbitrarily large) collections of them to a file for persistent storage. After that, the data can be retrieved and post-processed quite easily with PyTables or even with another HDF5 application (in C, Fortran, Java or whatever language that provides a library to interface with HDF5).
Other important entities in PyTables are array objects, which are analogous to tables with the difference that all of their components are homogeneous. They come in different flavors, like generic (they provide a quick and fast way to deal with for numerical arrays), enlargeable (arrays can be extended along a single dimension) and variable length (each row in the array can have a different number of elements).
The next section describes the most interesting capabilities of PyTables.
Main Features¶
PyTables takes advantage of the object orientation and introspection capabilities offered by Python, the powerful data management features of HDF5, and NumPy’s flexibility and Numexpr’s high-performance manipulation of large sets of objects organized in a grid-like fashion to provide these features:
Support for table entities: You can tailor your data adding or deleting records in your tables. Large numbers of rows (up to 2**63, much more than will fit into memory) are supported as well.
Multidimensional and nested table cells: You can declare a column to consist of values having any number of dimensions besides scalars, which is the only dimensionality allowed by the majority of relational databases. You can even declare columns that are made of other columns (of different types).
Indexing support for columns of tables: Very useful if you have large tables and you want to quickly look up for values in columns satisfying some criteria.
Support for numerical arrays: NumPy (see [NUMPY]) arrays can be used as a useful complement of tables to store homogeneous data.
Enlargeable arrays: You can add new elements to existing arrays on disk in any dimension you want (but only one). Besides, you are able to access just a slice of your datasets by using the powerful extended slicing mechanism, without need to load all your complete dataset in memory.
Variable length arrays: The number of elements in these arrays can vary from row to row. This provides a lot of flexibility when dealing with complex data.
Supports a hierarchical data model: Allows the user to clearly structure all data. PyTables builds up an object tree in memory that replicates the underlying file data structure. Access to objects in the file is achieved by walking through and manipulating this object tree. Besides, this object tree is built in a lazy way, for efficiency purposes.
User defined metadata: Besides supporting system metadata (like the number of rows of a table, shape, flavor, etc.) the user may specify arbitrary metadata (as for example, room temperature, or protocol for IP traffic that was collected) that complement the meaning of actual data.
Ability to read/modify generic HDF5 files: PyTables can access a wide range of objects in generic HDF5 files, like compound type datasets (that can be mapped to Table objects), homogeneous datasets (that can be mapped to Array objects) or variable length record datasets (that can be mapped to VLArray objects). Besides, if a dataset is not supported, it will be mapped to a special UnImplemented class (see The UnImplemented class), that will let the user see that the data is there, although it will be unreachable (still, you will be able to access the attributes and some metadata in the dataset). With that, PyTables probably can access and modify most of the HDF5 files out there.
Data compression: Supports data compression (using the Zlib, LZO, bzip2 and Blosc compression libraries) out of the box. This is important when you have repetitive data patterns and don’t want to spend time searching for an optimized way to store them (saving you time spent analyzing your data organization).
High performance I/O: On modern systems storing large amounts of data, tables and array objects can be read and written at a speed only limited by the performance of the underlying I/O subsystem. Moreover, if your data is compressible, even that limit is surmountable!
Support of files bigger than 2 GB: PyTables automatically inherits this capability from the underlying HDF5 library (assuming your platform supports the C long long integer, or, on Windows, __int64).
Architecture-independent: PyTables has been carefully coded (as HDF5 itself) with little-endian/big-endian byte ordering issues in mind. So, you can write a file on a big-endian machine (like a Sparc or MIPS) and read it on other little-endian machine (like an Intel or Alpha) without problems. In addition, it has been tested successfully with 64 bit platforms (Intel-64, AMD-64, PowerPC-G5, MIPS, UltraSparc) using code generated with 64 bit aware compilers.
The Object Tree¶
The hierarchical model of the underlying HDF5 library allows PyTables to manage tables and arrays in a tree-like structure. In order to achieve this, an object tree entity is dynamically created imitating the HDF5 structure on disk. The HDF5 objects are read by walking through this object tree. You can get a good picture of what kind of data is kept in the object by examining the metadata nodes.
The different nodes in the object tree are instances of PyTables classes. There are several types of classes, but the most important ones are the Node, Group and Leaf classes. All nodes in a PyTables tree are instances of the Node class. The Group and Leaf classes are descendants of Node. Group instances (referred to as groups from now on) are a grouping structure containing instances of zero or more groups or leaves, together with supplementary metadata. Leaf instances (referred to as leaves) are containers for actual data and can not contain further groups or leaves. The Table, Array, CArray, EArray, VLArray and UnImplemented classes are descendants of Leaf, and inherit all its properties.
Working with groups and leaves is similar in many ways to working with directories and files on a Unix filesystem, i.e. a node (file or directory) is always a child of one and only one group (directory), its parent group [1]. Inside of that group, the node is accessed by its name. As is the case with Unix directories and files, objects in the object tree are often referenced by giving their full (absolute) path names. In PyTables this full path can be specified either as string (such as ‘/subgroup2/table3’, using / as a parent/child separator) or as a complete object path written in a format known as the natural name schema (such as file.root.subgroup2.table3).
Support for natural naming is a key aspect of PyTables. It means that the names of instance variables of the node objects are the same as the names of its children [2]. This is very Pythonic and intuitive in many cases. Check the tutorial Reading (and selecting) data in a table for usage examples.
You should also be aware that not all the data present in a file is loaded into the object tree. The metadata (i.e. special data that describes the structure of the actual data) is loaded only when the user want to access to it (see later). Moreover, the actual data is not read until she request it (by calling a method on a particular node). Using the object tree (the metadata) you can retrieve information about the objects on disk such as table names, titles, column names, data types in columns, numbers of rows, or, in the case of arrays, their shapes, typecodes, etc. You can also search through the tree for specific kinds of data then read it and process it. In a certain sense, you can think of PyTables as a tool that applies the same introspection capabilities of Python objects to large amounts of data in persistent storage.
It is worth noting that PyTables sports a metadata cache system that loads nodes lazily (i.e. on-demand), and unloads nodes that have not been used for some time (following a Least Recently Used schema). It is important to stress out that the nodes enter the cache after they have been unreferenced (in the sense of Python reference counting), and that they can be revived (by referencing them again) directly from the cache without performing the de-serialization process from disk. This feature allows dealing with files with large hierarchies very quickly and with low memory consumption, while retaining all the powerful browsing capabilities of the previous implementation of the object tree. See [OPTIM] for more facts about the advantages introduced by this new metadata cache system.
To better understand the dynamic nature of this object tree entity, let’s start with a sample PyTables script (which you can find in examples/objecttree.py) to create an HDF5 file:
import tables as tb
class Particle(tb.IsDescription):
identity = tb.StringCol(itemsize=22, dflt=" ", pos=0) # character String
idnumber = tb.Int16Col(dflt=1, pos = 1) # short integer
speed = tb.Float32Col(dflt=1, pos = 2) # single-precision
# Open a file in "w"rite mode
fileh = tb.open_file("objecttree.h5", mode = "w")
# Get the HDF5 root group
root = fileh.root
# Create the groups
group1 = fileh.create_group(root, "group1")
group2 = fileh.create_group(root, "group2")
# Now, create an array in root group
array1 = fileh.create_array(root, "array1", ["string", "array"], "String array")
# Create 2 new tables in group1
table1 = fileh.create_table(group1, "table1", Particle)
table2 = fileh.create_table("/group2", "table2", Particle)
# Create the last table in group2
array2 = fileh.create_array("/group1", "array2", [1,2,3,4])
# Now, fill the tables
for table in (table1, table2):
# Get the record object associated with the table:
row = table.row
# Fill the table with 10 records
for i in range(10):
# First, assign the values to the Particle record
row['identity'] = f'This is particle: {i:2d}'
row['idnumber'] = i
row['speed'] = i * 2.
# This injects the Record values
row.append()
# Flush the table buffers
table.flush()
# Finally, close the file (this also will flush all the remaining buffers!)
fileh.close()
This small program creates a simple HDF5 file called objecttree.h5 with the structure that appears in Figure 1 [3]. When the file is created, the metadata in the object tree is updated in memory while the actual data is saved to disk. When you close the file the object tree is no longer available. However, when you reopen this file the object tree will be reconstructed in memory from the metadata on disk (this is done in a lazy way, in order to load only the objects that are required by the user), allowing you to work with it in exactly the same way as when you originally created it.
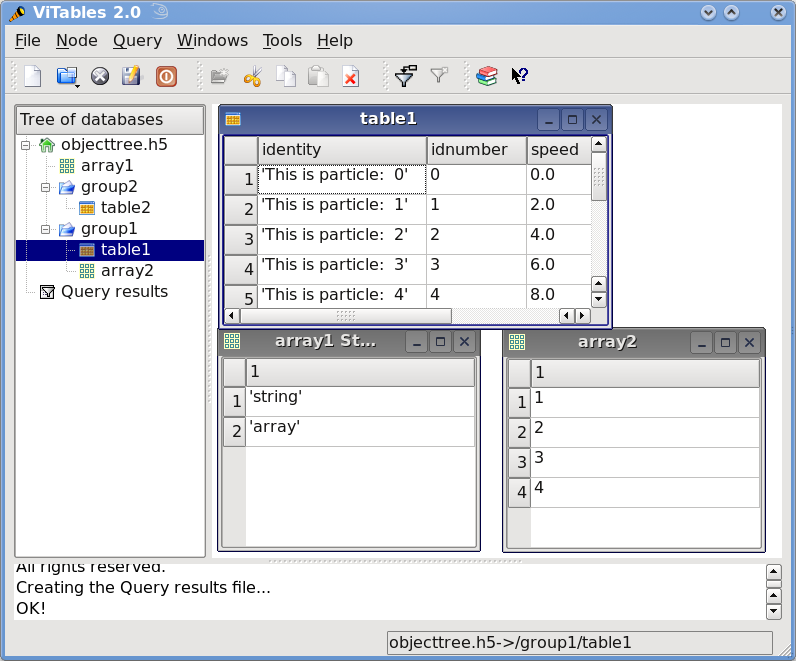
Figure 1: An HDF5 example with 2 subgroups, 2 tables and 1 array.¶
In Figure2, you can see an example of the object tree created when the above objecttree.h5 file is read (in fact, such an object tree is always created when reading any supported generic HDF5 file). It is worthwhile to take your time to understand it [4]. It will help you understand the relationships of in-memory PyTables objects.
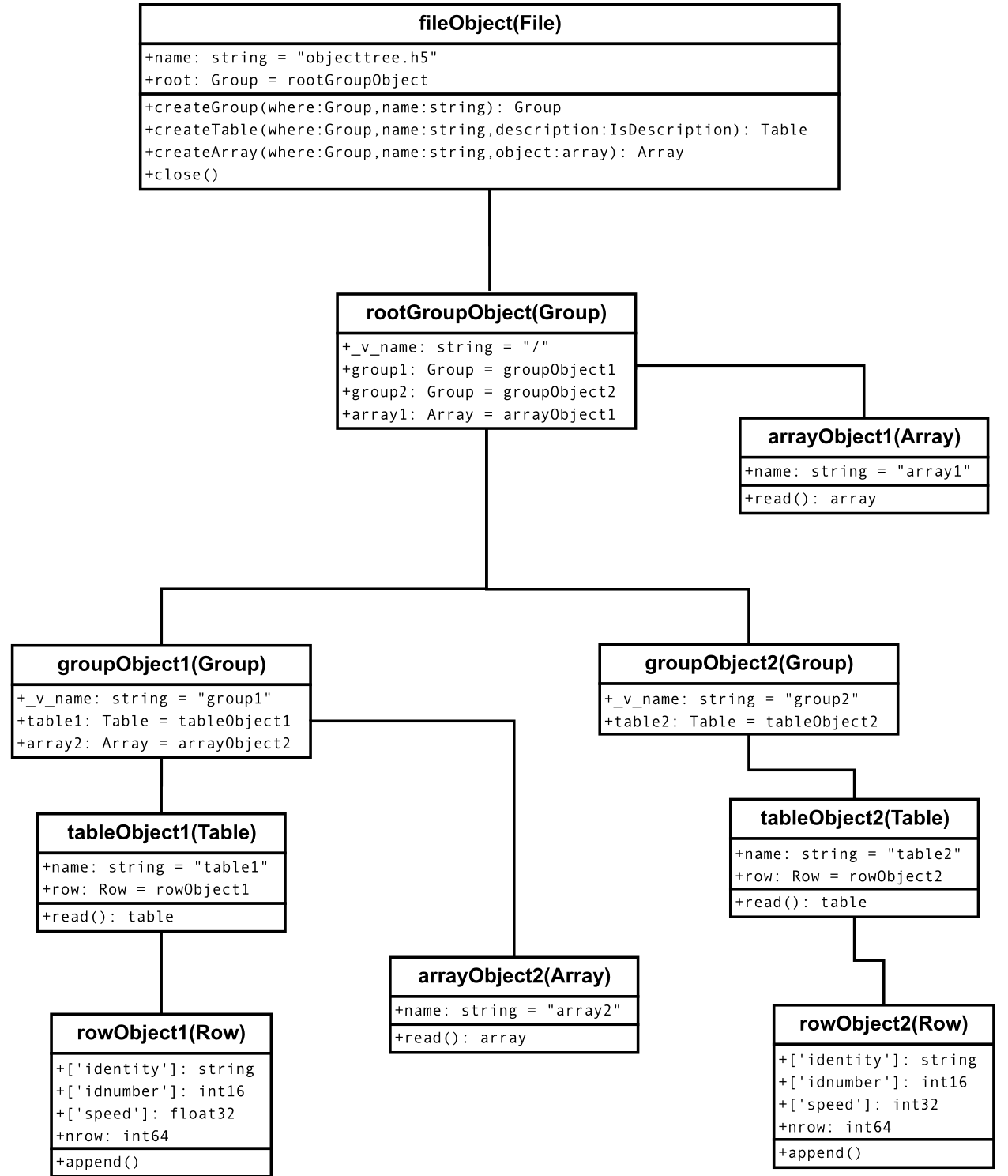
Figure 2: A PyTables object tree example.¶